Tutorial for OikoBase
Navigating the Facepage
The Face page is the entry point to OikoBase. There are 10 links to access the various features:
1. “About”- this link takes you to a brief description of Oikopleura and the OikoBase project, as well as funding sources that contributed to the construction of OikoBase.
2. “OikoBase Tutorial”- this link provides a comprehensive tutorial of OikoBase. We recommend reading through this document in its entirety in order to become familiar with the capabilities of this resource.
3. “People”- this link takes you to the people who contributed to OikoBase; clicking on the individual names takes you to a personal description of their involvement.
4. “GBrowse”- this link takes you to the genome browser of OikoBase (described below). Here is where you can access expression profiles of transcripts across the genome as well as information on the annotated genes.
5. “BLAST”- this link takes you to the BLAST function of OikoBase (described below). Several programs can be used to query four databases: 1) genomic sequence, 2) transcripts, 3) peptides, and, 4) ESTs.
6. “Methods”- This link takes you to a comprehensive description of the methods used. Included here are relevant animal culture and molecular biological procedures, in addition to microarray hybridization information, gene annotation and processing of the array data. A detailed description of pre-processing of raw microarray probe intensities is included (also see Figure 1 in Methods), along with a description of how TARs/superTARs are generated. A pipeline for processing of probe intensities to generate gene expression values (as well as expression values of superTARs) is shown in Figure 2 of Methods. Validation of gene expression profiles is provided in Figure 3 and Table 1 of Methods. Finally, gene name, Gene Ontology and InterPro domain annotation procedures are described.
7. “Gene Expression Matrix”- this link takes you to a webpage with a sortable table of expression values for all genes for all developmental time points. By clicking on any header link (light blue boxes at the top of each column), expression values will be sorted from high to low (the selected box should turn dark blue while the information is sorted). Re-clicking on the header link will sort expression values from low to high. All genes are represented by active links which will directly take you to the location of that gene in the genome browser (genes can also be sorted by their GSOIDG numbers). Please be patient after selecting the Gene Expression Matrix link to allow for complete loading of the table, which consists of expression values for 18,000+ genes at 18 different time points! Additionally, when selecting a header link, allow time for the data to sort.
8. “Downloads”- downloadable documents include a table of Gene Ontology terms for all genes; a table of InterPro terms for all genes; a GFF of the annotated genes; fasta files of the genome sequence of all scaffolds, the annotated gene transcripts, and the annotated gene peptides; and a compressed text file version of the Gene Expression Matrix.
9. “Related Links”- Links to all affiliated University departments and centers are included.
10. “Updates”- This section will detail future changes to OikoBase.
Navigating the Genome Browser
Clicking on the GBrowse link takes you to the Genome Browser function. The upper left window allows you to enter information to take you to a particular scaffold or gene location. Additionally, a scaffold can be directly selected from the Examples section. Alternatively, a gene (GSOIDG), transcript (GSOIDT), or protein (GSOIDP) ID can be entered into the box and searched, bringing you to the scaffold and specific location of the gene of interest. If you are interested in searching for particular gene terms, for example “gata-3” or “transcription factor”, you may do so using a wildcard symbol (*). Thus, searching for *gata-3* will retrieve any genes that have gata-3 as part of their name.
Scroll and zoom bars are located in the center of the screen, and a dropdown menu allows you to visualize different sized genomic regions. Another dropdown menu immediately above allows you to annotate restriction sites or download a fasta or sequence file of the scaffold that is being accessed. At the bottom of the screen, various tracks can be selected. Under the “General” heading, tracks for protein-coding portions of genes (CDS), GC content of the DNA (DNA), mRNAs of the annotated genes (mRNAs), and annotated genes (Named gene) are selectable. Under the “NimbleGen_Expression” heading, tracks for 18 different developmental transcriptome time points can be selected. To visualize the same data transformed into the log2 scale, the viewer can access the files under the “NimbleGen_Expression_log2” heading. Please note that log2 transformed data appear “smoothened” and less choppy, which may be preferable for some users who wish to view larger scale dynamic expression changes. Selecting the untransformed data files should be preferable for viewers who wish to identify more subtle changes in gene expression. Once the expression tracks are loaded, a variety of configuration features can be accessed using the “configure this track” button at the upper left of the track. Additionally, the track can be downloaded using the “download this track” button immediately to the right of the “configure this track” button.
To access additional information about any gene in the browser window, scroll the mouse arrow over the gene of interest. A bubble will appear that gives the GSOID gene number as well as a gene name if relevant (see Methods section for gene name annotation of genes). Clicking on the gene model will open a different bubble that gives you access to GO or InterPro terms (see Methods section for GO and InterPro annotations of genes) associated with that gene product. Transcript or peptide sequences can also be downloaded by clicking on the appropriate links in the bubble. An example of navigating the genome browser is shown in Figure 1:
Figure 1. Data mining in OikoBase. A) Browser window of a 50 kb segment of scaffold one showing several genes, with the tailshift expression track selected as an example of a time point. B) By placing the pointer over a gene model (1) (here, the 6th gene from the left) a balloon appears that specifies the gene name, gata-3 (if it is homologous to a known named gene) and its genomic coordinates (2). C) By clicking on the gene model, a different balloon (3) is opened in its place which allows links to both the Gene Ontology website (4) and the InterPro website. By clicking on either link, the browser is redirected to the relevant GO or InterPro terms associated with this gene which catalogue information regarding biological processes, molecular functions and cellular components as well as evolutionarily conserved protein domains. Options to directly download nucleotide (TRANSCRIPT SEQ) and protein (PEPTIDE SEQ) are also provided in this balloon.
Performing a BLAST Search
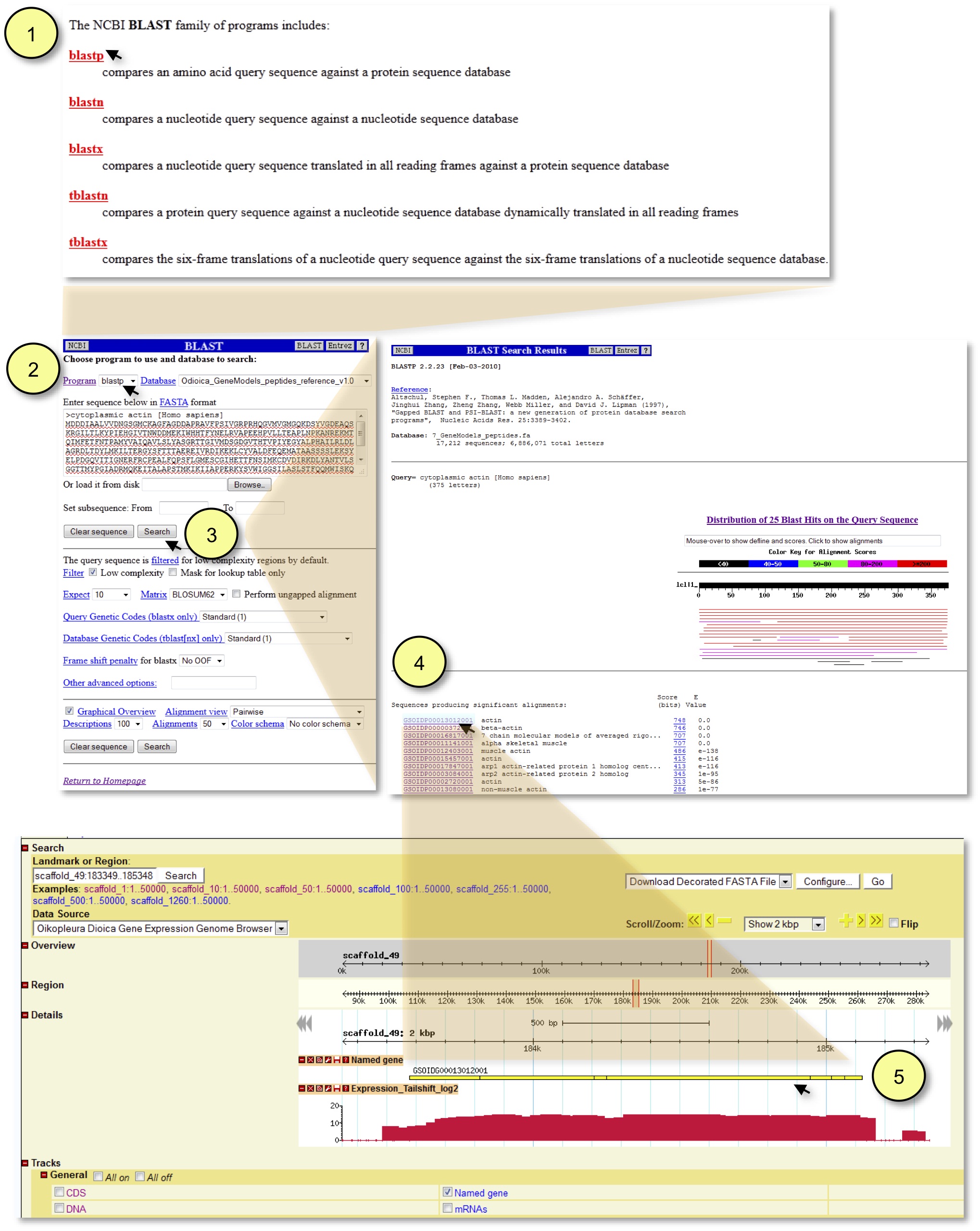
To perform a BLAST search, access the “Programs available for the BLAST search” page using the Facepage BLAST link. From this page, clicking on either “blastp” or “blastx” will take you to the BLAST function which has automatically linked either program with the Gene Model Peptides database. Clicking on blastn, tblastn, or tblastx will additionally require selection of the appropriate database to be searched, since these BLAST functions can be used to search the transcripts, ESTs, or genome assembly databases. As a default, all three links automatically select the transcripts database. The standard BLAST parameters can be adjusted at the bottom of the page. Once the desired settings are in place, you can enter your sequence of interest and click on the search button. This will take you to a results page with a list of active links representing all hits. Links associated with transcripts or proteins consist of the transcript (GSOIDT) or protein (GSOIDP) ID followed by the gene name, if one exists. Clicking on one of the links will take you to the relevant scaffold and gene location, where you can then access the developmental transcriptome data for that gene for as many times points as you wish to select. BLAST searches of DNA sequences will generate alignments to particular scaffolds; the active links will take you directly to the relevant scaffold and start position of the alignment + 1000 bp. The zoom features can then be used to generate the desired view of the relevant genomic regions.
BLAST searches of DNA sequences will generate alignments to particular scaffolds; the active links will take you directly to the relevant scaffold and start position of the alignment + 1000 bp. The zoom features can then be used to generate the desired view of the relevant genomic regions.
An example of performing a BLAST search is shown in Figure 2:
Figure 2. Obtaining developmental expression profiles for O. dioica homologs of genes-of-interest from other organisms. To provide an example of how to use this pipeline, the human cytoplasmic actin protein has been selected as the query sequence.First, the “Programs available for the BLAST search” page (1) is accessed by clicking the Facepage BLAST link. The desired Program should then be matched up with the appropriate Database using the dropdown menus (2). Note that selecting “blastp” (as in this example) or “blastx” will automatically pair these programs with the “O. dioica peptides reference” database. After executing the search (3), the desired putative O. dioica homologs are displayed on the BLAST search results page where unique GSOID identifiers and corresponding gene names are provided (4). By clicking on the desired live link GSOIDP identifier the user is taken to the location of the putative homolog (5). Subsequently, any desired additional annotation or transcript tracks can be activated in the browser. Here, the expression of an O. dioica cytoplasmic actin homolog is shown at the tailshift stage with the view zoomed to 2 kb.
Return to Homepage
Each page in OikoBase has a Return to Homepage link at the bottom of the page for convenience.
Contacting OikoBase/Help Request
To contact OikoBase for additional assistance, please send an email to john-manak@uiowa.edu or mrutyunjaya-parida@uiowa.edu